ShapeFactor#
[1]:
import logging
import json
import numpy as np
import matplotlib.pyplot as plt
import pyhf
from pyhf.contrib.viz import brazil
logging.basicConfig(level=logging.INFO)
[2]:
def prep_data(sourcedata):
spec = {
'channels': [
{
'name': 'signal',
'samples': [
{
'name': 'signal',
'data': sourcedata['signal']['bindata']['sig'],
'modifiers': [
{'name': 'mu', 'type': 'normfactor', 'data': None}
],
},
{
'name': 'bkg1',
'data': sourcedata['signal']['bindata']['bkg1'],
'modifiers': [
{
'name': 'coupled_shapefactor',
'type': 'shapefactor',
'data': None,
}
],
},
],
},
{
'name': 'control',
'samples': [
{
'name': 'background',
'data': sourcedata['control']['bindata']['bkg1'],
'modifiers': [
{
'name': 'coupled_shapefactor',
'type': 'shapefactor',
'data': None,
}
],
}
],
},
]
}
pdf = pyhf.Model(spec, poi_name="mu")
data = []
for channel in pdf.config.channels:
data += sourcedata[channel]['bindata']['data']
data = data + pdf.config.auxdata
return data, pdf
[3]:
source = {
"channels": {
"signal": {
"binning": [2, -0.5, 1.5],
"bindata": {
"data": [220.0, 230.0],
"bkg1": [100.0, 70.0],
"sig": [20.0, 20.0],
},
},
"control": {
"binning": [2, -0.5, 1.5],
"bindata": {"data": [200.0, 300.0], "bkg1": [100.0, 100.0]},
},
}
}
data, pdf = prep_data(source['channels'])
print(f'data: {data}')
init_pars = pdf.config.suggested_init()
print(f'expected data: {pdf.expected_data(init_pars)}')
par_bounds = pdf.config.suggested_bounds()
INFO:pyhf.pdf:Validating spec against schema: model.json
INFO:pyhf.pdf:adding modifier mu (1 new nuisance parameters)
INFO:pyhf.pdf:adding modifier coupled_shapefactor (2 new nuisance parameters)
data: [200.0, 300.0, 220.0, 230.0]
expected data: [100. 100. 120. 90.]
[4]:
print(f'initialization parameters: {pdf.config.suggested_init()}')
unconpars = pyhf.infer.mle.fit(data, pdf)
print(f'parameters post unconstrained fit: {unconpars}')
initialization parameters: [1.0, 1.0, 1.0]
parameters post unconstrained fit: [1.00004623 1.99998941 3.00000438]
/srv/conda/envs/notebook/lib/python3.7/site-packages/pyhf/tensor/numpy_backend.py:334: RuntimeWarning: divide by zero encountered in log
return n * np.log(lam) - lam - gammaln(n + 1.0)
[5]:
(
obs_limit,
exp_limits,
(poi_tests, tests),
) = pyhf.infer.intervals.upper_limits.upper_limit(
data, pdf, np.linspace(0, 5, 61), level=0.05, return_results=True
)
/srv/conda/envs/notebook/lib/python3.7/site-packages/pyhf/infer/calculators.py:352: RuntimeWarning: invalid value encountered in double_scalars
teststat = (qmu - qmu_A) / (2 * self.sqrtqmuA_v)
[6]:
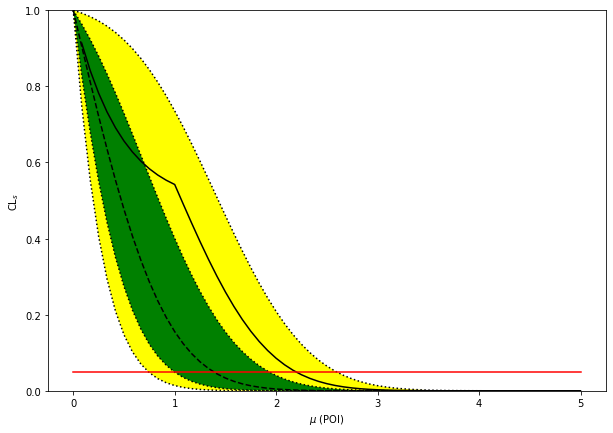
fig, ax = plt.subplots(figsize=(10, 7))
artists = brazil.plot_results(poi_tests, tests, test_size=0.05, ax=ax)
print(f'expected upper limits: {exp_limits}')
print(f'observed upper limit : {obs_limit}')
expected upper limits: [array(0.74138115), array(0.994935), array(1.38451391), array(1.92899382), array(2.59407668)]
observed upper limit : 2.1945969322493744