Multibin Coupled HistoSys#
[1]:
import json
import logging
import matplotlib.pyplot as plt
import numpy as np
import pyhf
from pyhf import Model
from pyhf.contrib.viz import brazil
logging.basicConfig(level=logging.INFO)
[2]:
def prep_data(sourcedata):
spec = {
"channels": [
{
"name": "signal",
"samples": [
{
"name": "signal",
"data": sourcedata["signal"]["bindata"]["sig"],
"modifiers": [
{"name": "mu", "type": "normfactor", "data": None}
],
},
{
"name": "bkg1",
"data": sourcedata["signal"]["bindata"]["bkg1"],
"modifiers": [
{
"name": "coupled_histosys",
"type": "histosys",
"data": {
"lo_data": sourcedata["signal"]["bindata"][
"bkg1_dn"
],
"hi_data": sourcedata["signal"]["bindata"][
"bkg1_up"
],
},
}
],
},
{
"name": "bkg2",
"data": sourcedata["signal"]["bindata"]["bkg2"],
"modifiers": [
{
"name": "coupled_histosys",
"type": "histosys",
"data": {
"lo_data": sourcedata["signal"]["bindata"][
"bkg2_dn"
],
"hi_data": sourcedata["signal"]["bindata"][
"bkg2_up"
],
},
}
],
},
],
},
{
"name": "control",
"samples": [
{
"name": "background",
"data": sourcedata["control"]["bindata"]["bkg1"],
"modifiers": [
{
"name": "coupled_histosys",
"type": "histosys",
"data": {
"lo_data": sourcedata["control"]["bindata"][
"bkg1_dn"
],
"hi_data": sourcedata["control"]["bindata"][
"bkg1_up"
],
},
}
],
}
],
},
]
}
pdf = Model(spec, poi_name="mu")
data = []
for channel in pdf.config.channels:
data += sourcedata[channel]["bindata"]["data"]
data = data + pdf.config.auxdata
return data, pdf
[3]:
validation_datadir = "../../../validation/data"
[4]:
with open(
validation_datadir + "/2bin_2channel_coupledhisto.json", encoding="utf-8"
) as spec:
source = json.load(spec)
data, pdf = prep_data(source["channels"])
print(f"data: {data}")
init_pars = pdf.config.suggested_init()
par_bounds = pdf.config.suggested_bounds()
unconpars = pyhf.infer.mle.fit(data, pdf, init_pars, par_bounds)
print(f"parameters post unconstrained fit: {unconpars}")
conpars = pyhf.infer.mle.fixed_poi_fit(0.0, data, pdf, init_pars, par_bounds)
print(f"parameters post constrained fit: {conpars}")
pdf.expected_data(conpars)
INFO:pyhf.pdf:Validating spec against schema: model.json
INFO:pyhf.pdf:adding modifier coupled_histosys (1 new nuisance parameters)
INFO:pyhf.pdf:adding modifier mu (1 new nuisance parameters)
data: [110.0, 105.0, 170.0, 220.0, 0.0]
parameters post unconstrained fit: [-0.30257894 0.63607078]
parameters post constrained fit: [0.29087602 0. ]
[4]:
array([106.80676913, 104.01075137, 154.36314037, 213.86871072,
0.29087602])
[5]:
def invert_interval(test_mus, cls_obs, cls_exp, test_size=0.05):
crossing_test_stats = {"exp": [], "obs": None}
for cls_exp_sigma in cls_exp:
crossing_test_stats["exp"].append(
np.interp(
test_size, list(reversed(cls_exp_sigma)), list(reversed(test_mus))
)
)
crossing_test_stats["obs"] = np.interp(
test_size, list(reversed(cls_obs)), list(reversed(test_mus))
)
return crossing_test_stats
[6]:
test_pois = np.linspace(0, 5, 61)
results = [
pyhf.infer.hypotest(
test_poi, data, pdf, init_pars, par_bounds, return_expected_set=True
)
for test_poi in test_pois
]
cls_obs = np.array([result[0] for result in results]).flatten()
cls_exp = [np.array([result[1][i] for result in results]).flatten() for i in range(5)]
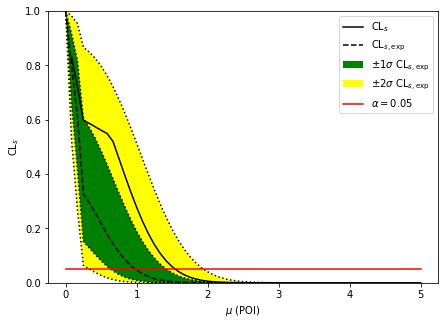
[7]:
fig, ax = plt.subplots()
fig.set_size_inches(7, 5)
artists = brazil.plot_results(test_pois, results, ax=ax)
invert_interval(test_pois, cls_obs, cls_exp)
[7]:
{'exp': [0.3379557904310767,
0.6355433028074602,
0.980748685945719,
1.4196566943996904,
1.9446272307776151],
'obs': 1.5452056912523955}